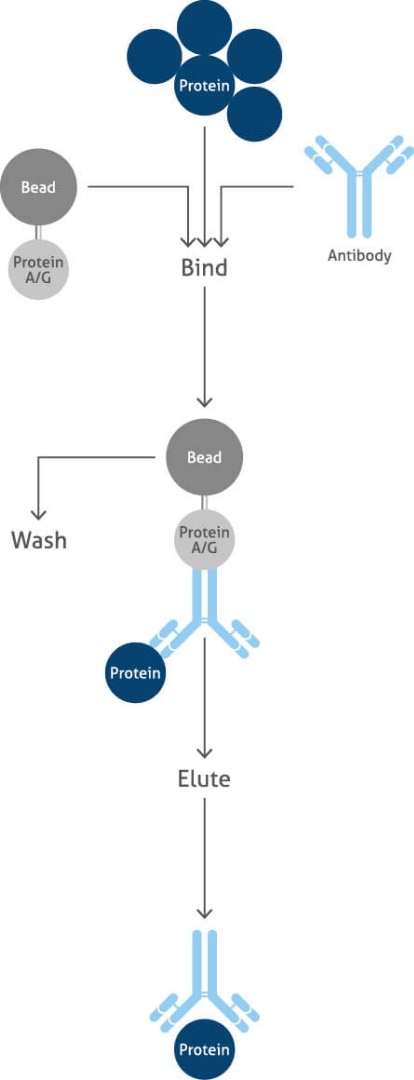
Immunoprecipitation
Immunoprecipitation (IP) is a precipitation technique that purifies and enriches a protein of interest, allowing the identification of protein-protein interactions in proteomics workflows. IP is an important technique used to investigate the presence, relative abundance, size, upregulation or downregulation, stability, post-translational modifications (PTMs), and interactions between proteins.
Antibodies for endogenous targets
Explore polyclonal or monoclonal primary antibodies for the IP of untagged, endogenous proteins of interest.
Non visible textNanobodies for proteins and peptide tags
Explore ChromoTek’s Nano-Traps for the IP of proteins carrying protein tags like GFP, mCherry or GST or carrying epitope tags like Spot-tag®, Myc-tag or V5-tag.
Non visible textWestern blot after IP
Explore WB products for the detection of the precipitated protein of interest and its interaction partners:
- Over 13,000 polyclonal and monoclonal primary antibodies against the protein of interest
- Tag antibodies against a protein or peptide tag
- Secondary antibodies
- WB accessories [SignalBright, Protein ladders]
Isotype control antibodies
Explore isotype control antibodies to find the right negative control antibody for your IP antibody to ensure highly specific results.
Non visible textWatch why GFP-Trap gives the best results for immunoprecipitation (IP)
Low background, no extra bands & high specificity will improve your pulldown assay significantly. Alpaca Alice shows how it works. Visit www.ptglab.com/nano-trap-free-sample/ to order a free test sample.
Featured Products
GFP-Trap® Agarose
| Specificity | GFP, eGFP, CFP, YFP, BFP |
| Applications | IP, CoIP, ChIP, RIP |
Myc-Trap® Agarose
| Specificity | Myc-Tag |
| Applications | IP, CoIP, ChIP, RIP |
HA tag Polyclonal antibody
| Reactivity | Recombinant Protein, Human, Mouse, Plant, Yeast |
| Applications | WB, IP, IHC, IF, CoIP, ChIP, ELISA |
Rabbit IgG control Polyclonal antibody
| Reactivity | Rabbit, Human, Mouse, Plasmodium Falciparum, Rat |
| Applications | WB, RIP, IP, IHC, IF, FC, ChIP, ELISA |
Resources
Videos
Immunoprecipitation Videos
Short technical tips videos on how to optimize your IP experiments
Non visible textProtocols
Immunoprecipitation Guide
Protocols and tips for optimizing your IP experiments
Non visible textBlog
Optimize your western blot protocol
7 Tips For Optimizing Your Western Blotting Experiments.
Non visible textVideos
Immunoprecipitation Videos
Short technical tips videos on how to optimize your IP experiments
Non visible textProtocols
Immunoprecipitation Guide
Protocols and tips for optimizing your IP experiments
Non visible textBlog
Optimize your western blot protocol
7 Tips For Optimizing Your Western Blotting Experiments.
Non visible text